Wanted: a perfect reproducible paper workflow
Last week I submitted my first article as part of my PhD. This paper was written in R markdown and is fully reproducible. It means that a single document contains all the manuscript but also all the code for the analysis and figure generation. All the pre-processings are also reproducible and the code is available on my gitlab. I am pretty happy to contribute to reproducible science, and I hope that my code is user-friendly enough to enable other people using it. Truth is, it was not the most straightforward way to write the paper, and consumed a lot of time. It does not mean it wasn’t worth it, and that I would not do it again. However, I would like to improve this reproducible workflow and learn from other people’s experience. That is why I am writing this short post to talk about the workflow we adopted and the challenges associated with this workflow, and start to think about how we could do differently in the future.
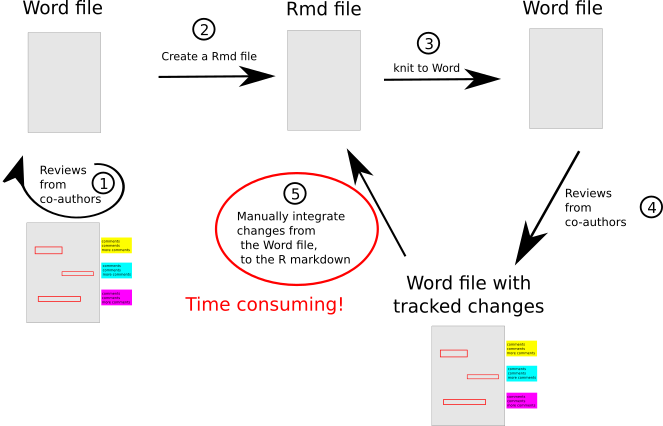
What is the workflow we adopted?
On the figure above, I schematized the workflow we adopted for writing this paper. First, we iterated only on a word document (step 1) to be able to track the changes made by my co-authors. This allowed to have my sentences corrected and to improve my writing. After the draft was relatively mature, I created a R markdown (Rmd) file using the text we had come up with, together with all the code for the analysis and figure generation (step 2). From this point, the internal reviewing process worked as follows: I knited the Rmd to Word (step 3), then my co-authors would successively add changes and comment the draft (step 4). When they were all done, I then manually included their changes back in the Rmd file, mostly by overusing the Ctrl + C and Ctrl + V commands (step 3). This was very time consuming. For each edit: I spotted the edit in word, copied the sentence, then I searched the exact location of the corresponding text in R markdown (Screenshot below), then paste it there. Then, in order not to loose track of the changes I already had included, I accepted the changes in the word document.
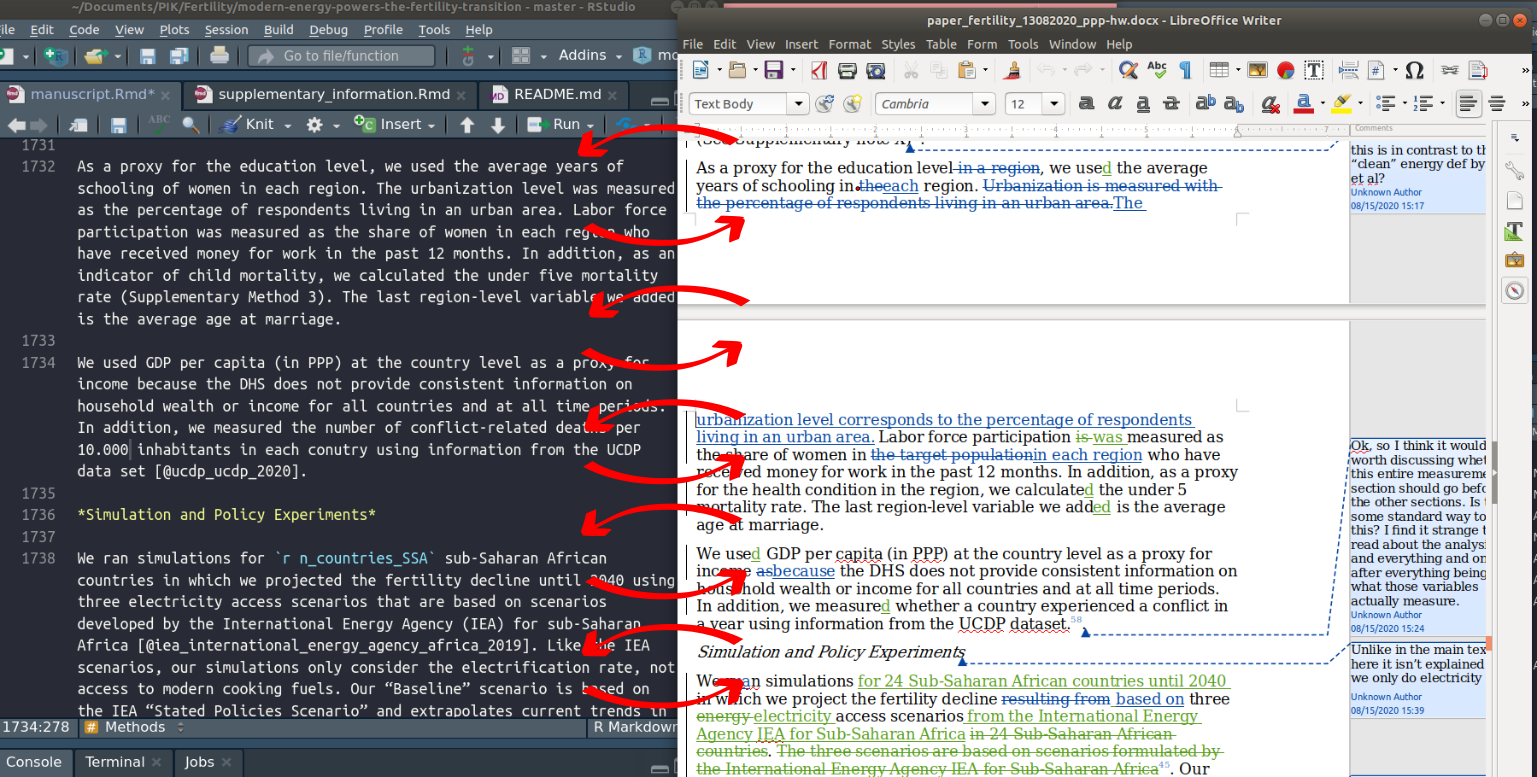
I repeated that for each changes, and for each the rounds of review (i.e. a lot of times, and a lot of headaches 🤯).

It was probably not the most intelligent/efficient way of proceeding, but once launched in the process, it was just easier to continue like this.
What are alternative workflows?
-
If track change is an absolute necessity (I tend to think so), I think the most obvious alternative is to use Word for the whole process, and to switch to R markdown at the end. However, it is hard to know when is actually going to be the last round of edit and getting used to the Rmarkdown-word process (steps 3-4) can take some time. If the paper needs to be submitted before a deadline, this could become quite stressful.
-
Another solution is that co-authors add changes directly on the Rmd. This requires that all co-authors are comfortable with navigating in a Rmd document, with potentially a lot of code chunks. Rstudio will soon release [Visual Rstudio](https://blog.rstudio.com/2020/09/30/rstudio-v1-4-preview-visual-markdown-editing/), that provides a user friendly visual text editor and this will hopefully attract more people to R markdown (and reproducibility 😊!). For example, users can now see their content change in real-time as they write. It also highlights spelling mistakes as in Word/Libre Office writer, really nice! You can try the preview [here](https://rstudio.com/products/rstudio/download/preview/). Unfortunately, track change or comments are still not available in this version. It makes it difficult to keep track of changes in collaborative work. One solution would be to commit to a git repository and visualizing the difference between two files, but this seems like an overly complicated process.
-
Using one Word document to write the paper, and have a separate Rmd file containing all the analysis and figure generation in a Rmd file. Then, whenever the numbers/tables/figures in the text needed to be updated, I would include them manually in the Word file.
-
I am aware that some R packages are being developed like redoc and reviewer. Unfortunately, the development of the former has now been suspended. Regarding reviewer, I have not tried it. My feeling is that people reluctant to using R or R markdown would have difficulty engaging with the method they propose. If you have a good experience with it, I would be happy to hear your feedback.
However I find that all of these workflow are imperfect for me, and I am still looking in search… If you have alternative solutions, please let me know!
Bottom line
The tools used to write a reproducible quantitative paper depend on your co-authors' respective tastes and their familiarity with available tools. The best workflow for you is not necessarily the best for your colleagues, and the other way around. One has to adapt and find a common ground, and also probably experience different workflows to see what fits best, under which circumstances. I am still looking for the perfect reproducible workflow, given the constrains that most of my colleagues are not necessarily familiar with writing in R markdown, and that I value track changes as part of the writing process.
What about you? I would be interested to hear about experience from people who have written reproducible papers. What was your workflow? Was it efficient? What were your constraints? Would you recommend it? Please share your story! ☺️